A growing body of research has documented the utility of magnetic resonance imaging technologies to support in vivo examination of brain organization and function in the nonhuman primate. Recent efforts have not only demonstrated the ability to recapitulate the findings of postmortem examinations, but also to provide novel insights into the organizational principles of nonhuman primate connectome and the functional interactions within it. These gains could only be made through in vivo studies. These advances are timely given the growing prominence of translational and comparative neuroscience studies in large-scale national and international initiatives focused on advancing our understanding of human brain function and ability to generate novel therapeutics for neurology and psychiatry.
Despite demonstrable feasibility and utility, the field of nonhuman primate imaging is only its infancy. Numerous challenges related to the acquisition and processing are still in the process of being worked out, and the bounds of what can be accomplished are only beginning to be explored. In this regard, the overarching goal of PRIMatE Data Exchange (PRIME-DE) is to create an open science resource for the neuroimaging community that will facilitate the mapping of the non-human primate connectome. To accomplish this, we will aggregate a combination of functional, diffusion and morphometric magnetic resonance imaging (MRI) datasets across laboratories around the world, and share the data with the larger scientific community.
The various webpages on the present site are intended to be informative regarding the PRIMatE Data Exchange (PRIME-DE). However, prior to data usage, we strongly recommend that you download and read the PRIMatE Data Exchange Data Descriptor.
Credits
- DirectorsDaniel Margulies1, Michael Milham2,3, Charlie Schroeder3,4, Christopher I. Petkov5, Sabine Kastner6
- Project ManagementMichael Milham2,3
- Data organizationNathalia Bianchini Esper2, Ting Xu2,3, Alexandre Franco 2,3
- LogoStacey Ascher2
- Centre National de la Recherche Scientifique (CNRS) UMR 7225, Institut du Cerveau et de la Moelle épinière, Paris, France
- Center for the Developing Brain, Child Mind Institute, New York, New York, United States of America
- Center for Biomedical Imaging and Neuromodulation at the Nathan S. Kline Institute for Psychiatric Research, Orangeburg, New York, United States of America
- Departments of Neurosurgery and Psychiatry, Columbia University College of Physicians and Surgeons, New York, New York, the United States of America
- Institute of Neuroscience, Newcastle University, Newcastle upon Tyne NE1 7RU, UK
- Princeton Neuroscience Institute, Princeton University, Princeton, the United States of America
Funding
Primary support for the work by Michael P. Milham and the INDI team was provided by gifts from Joseph P. Healy to the Child Mind Institute, as well as by the BRAIN Initiative (R01MH111439). MPM is a Randolph Cowen and Phyllis Green Scholar.
Primary support for the work by Charles Schroeder is provided by the BRAIN Initiative (R01MH111439) and the Sylvio O. Conte Center “Neurobiology and Dynamics of Active Sensing” (P50MH109429).
Primary support for the work by Daniel Margulies is provided by the Max Planck Society.
Data Collections
| Study Site | Investigators | Species | Subjects | Scanner | State | Structural T1 | Structural T2 | Resting State fMRI | Naturalistic Viewing fMRI | Task fMRI | Fieldmap | Diffusion MRI | Magnetic Resonance Spectroscopy | Processed Data | Template Provided | Usage Agreement |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aix-Marseille Université | Brochier Sein |
Macaca mulatta | 4 | Siemens Prisma 3T | Anesthetized | ✔ | ✔ | ✔ | CC-BY-NC-SA | |||||||
| California Institute of Technology | Rejimehr Tsao |
Macaca mulatta | 2 | Siemens TIM Trio 3T | Awake | 96 min | CC-BY-NC-SA | |||||||||
| East China Normal University - Chen | Chen | Macaca mulatta | 10 | Siemens TIM Trio 3T | Anesthetized | ✔ | CC-BY-NC-SA | |||||||||
| East China Normal University - Kwok | Kowk Zhou |
Macaca mulatta | 4 | Siemens TIM Trio 3T | Anesthetized | ✔ | ✔ | 8 min | ✔ | ✔ | CC-BY-NC-SA | |||||
| Georgetown University | Archakov DeWitt Kuśmierek Ortiz-Rios VanMeter Sams Jääskeläinen Rauschecker |
Macaca mulatta | 2 | Anesthetized | ✔ | ✔ | ✔ | CC-BY-NC-SA | ||||||||
| Institute of Neuroscience | Wang | Macaca mulatta, Macaca fascicularis | 8 | Siemens TIM Trio 3T | Anesthetized | ✔ | 20-40 min | ✔ | CC-BY-NC-SA | |||||||
| Institut des Sciences Cognitives Marc Jeannerod | Ben Hamed Hiba |
Macaca mulatta, Macaca fascicularis | 8 | Siemens Sonata 1.5T Siemens Prisma 3T |
Anesthetized/Awake | ✔ | ✔ | CC-BY-NC-SA | ||||||||
| Lyon Neuroscience Research Center | Hadj-Bouziane Meunier Guedj |
Macaca mulatta | 4 | Siemens Sonata 1.5T Siemens Prisma 3T |
Awake | ✔ | ✔ | ✔ | CC-BY-NC-SA | |||||||
| MADI@700, Bordeaux University | Caron Corbin Boissenin Gupta Nguyen Anandra Miraux Descoteaux Petit Wagner |
Macaca fascicularis | 9 | Siemens Prisma 3T | Anesthetized | ✔ | ✔ | ✔ | ✔ | ✔ | CC-BY | |||||
| McGill University | Mok Rudko Shmuel |
Macaca mulatta | 3 | Siemens TIM Trio 3T | Anesthetized | ✔ | ✔ | CC-BY-NC-SA | ||||||||
| Mount Sinai School of Medicine | Croxson Fleysher |
Macaca mulatta | 8 | Philips Achieva 3T | Anesthetized | ✔ | ✔ | 43 min | ✔ | ✔ | CC-BY-NC-SA | |||||
| Mount Sinai School of Medicine | Croxson Fleysher Froudist-Walsh Damatac Nagy |
Macaca mulatta | 5 | Siemens Skyra 3T | Anesthetized | ✔ | ✔ | ✔ | CC-BY-NC-SA | |||||||
| Nathan Kline Institute | Schroeder Milham |
Macaca mulatta | 2 | Siemens TIM Trio 3T | Anesthetized/Awake | ✔ | 76-155 min | 55-345 min | CC-BY-NC-SA | |||||||
| National Institute of Mental Health | Leopold Russ |
Macaca mulatta | 3 | Bruker BioSpec Vertical 4.7T | Awake | ✔ | ✔ | 30-150 min | 170 min | DUA | ||||||
| National Institute of Mental Health | Messinger Jung Seidlitz Ungerleider |
Macaca mulatta | 3 | Bruker BioSpec Vertical 4.7T | Anesthetized/Awake | ✔ | 10-15 min | Cortical Thickness | ✔ | DUA | ||||||
| Netherlands Institute for Neuroscience | Klink Roelfsema |
Macaca mulatta | 2 | Philips Ingenia 3T | Anesthetized | ✔ | ✔ | 9.7 min | CC-BY-NC-SA | |||||||
| NeuroSpin | Jarraya Dehaene |
Macaca mulatta | 3 | Siemens TIM Trio 3T Siemens Prisma Fit |
Anesthetized | ✔ | ✔ | ✔ | CC-BY-NC-SA | |||||||
| Newcastle University | Petkov Nacef Thiele Poirier Balezeau Griffiths Schmid Rios |
Macaca mulatta | 14 | Bruker Vertical 4.7T | Anesthetized/Awake | ✔ | ✔ | 21.6 min | CC-BY-NC-SA | |||||||
| Oregon Health and Science University | Sullivan Fair |
Macaca mulatta | 2 | Siemens TIM Trio 3T | Anesthetized | ✔ | ✔ | 480 min | ✔ | CC-BY-NC-SA | ||||||
| Princeton University | Kastner Pinsk |
Macaca mulatta | 2 | Siemens Prisma VE11C 3T | Anesthetized | ✔ | ✔ | 60 min | ✔ | ✔ | CC-BY-NC-SA | |||||
| Rockefeller University | Schwiedrzik Freiwald Zarco |
Macaca mulatta, Macaca fascicularis | 6 | Siemens TIM Trio 3T | Anesthetized | ✔ | 80 min | ✔ | CC-BY-NC-SA | |||||||
| Stem Cell and Brain Research Institute | Procyk Wilson Amiez |
Macaca mulatta, Macaca fascicularis | 22 | Siemens Sonata 1.5T Siemens Prisma 3T |
Anesthetized | ✔ | ✔ | ✔ | CC-BY-NC-SA | |||||||
| Université de Lyon - CarMeN Laboratory | Canet-Soulas Chalet Becker |
Macaca Fascicularis | 20 | Biograph mMR PET-MRI 3T | Anesthetized | ✔ | ✔ | ✔ | DUA | |||||||
| University of California, Davis | Baxter Croxson Morrison |
Macaca mulatta | 19 | Siemens Skyra 3T | Anesthetized | ✔ | ✔ | 13.5 min | ✔ | ✔ | CC-BY-NC-SA | |||||
| University of California, Davis | Amaral Bennett |
Macaca mulatta | 56 | Siemens Tim Trio 3T GE Genesis Signa 1.5T |
Anesthetized | ✔ | ✔ | ✔ | ✔ | CC-BY-NC-SA | ||||||
| University of Minnesota | Yacoub Harel |
Macaca mulatta | 2 | Siemens Syngo B17 7T | Anesthetized | ✔ | 27 min | ✔ | ✔ | CC-BY-NC-SA | ||||||
| University of Oxford | Sallet Mars Rushworth |
Macaca mulatta | 20 | [Undusclosed] 3T | Anesthetized | ✔ | 53.43 min | CC-BY-NC-SA (See page for additional details) |
||||||||
| University of Oxford (PM) | Mars Sallet |
Macaca mulatta | 6 | Agilent 7T | Post-mortem | ✔ | CC-BY-NC-SA | |||||||||
| University of Rochester Medical Center | Fudge Kwok Sharma |
Macaque fascicularis | 3 | Siemens Prisma 3T | Anesthetized | ✔ | ✔ | ✔ | CC-BY-NC-SA | |||||||
| NIN Primate Brain Bank/Utrecht University | Navarrete Blezer Todorov Lindenfors Laland Reader |
See site page | 51 | Varian small-bore 9.4T Siemens Magnetom Trio 3T |
Post-mortem | ✔ | CC-BY-NC-SA | |||||||||
| University of Western Ontario | Everling Menon |
Macaca mulatta | 12 | Siemens Magnetom 7T | Anesthetized | ✔ | ✔ | 60 min | CC-BY-NC-SA | |||||||
| University of Wisconsin–Madison | Kalin Fox Oler Brin Tromp Alexander |
Macaca mulatta | 592 | GE Discovery_MR750 3T GE Signa EXCITE 3T |
Anesthetized | ✔ | 16 min | ✔ | ✔ | CC-BY-NC-SA |
Quality Assurance Protocol
Consistent with recent major FCP/INDI data releases, we made use of the Preprocessed Connectome Project Quality Assurance Protocol (QAP; Shehzad et al., 2013) to assess data quality for core MRI data modalities (i.e., functional MRI, morphometry MRI and diffusion MRI). More information about the quality control process used for HBN MRI data can be found at the PCP Quality Assurance Protocol webpage.
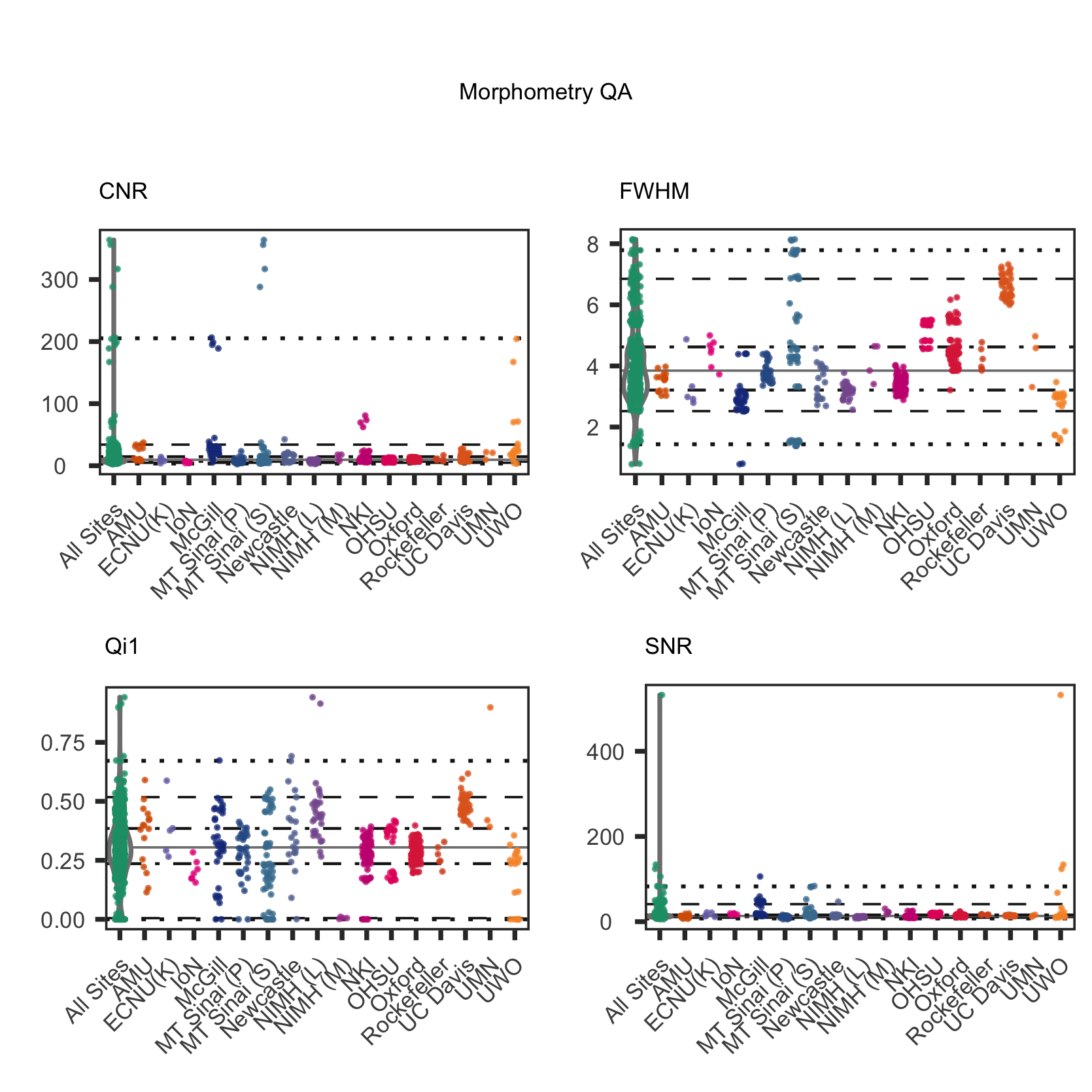
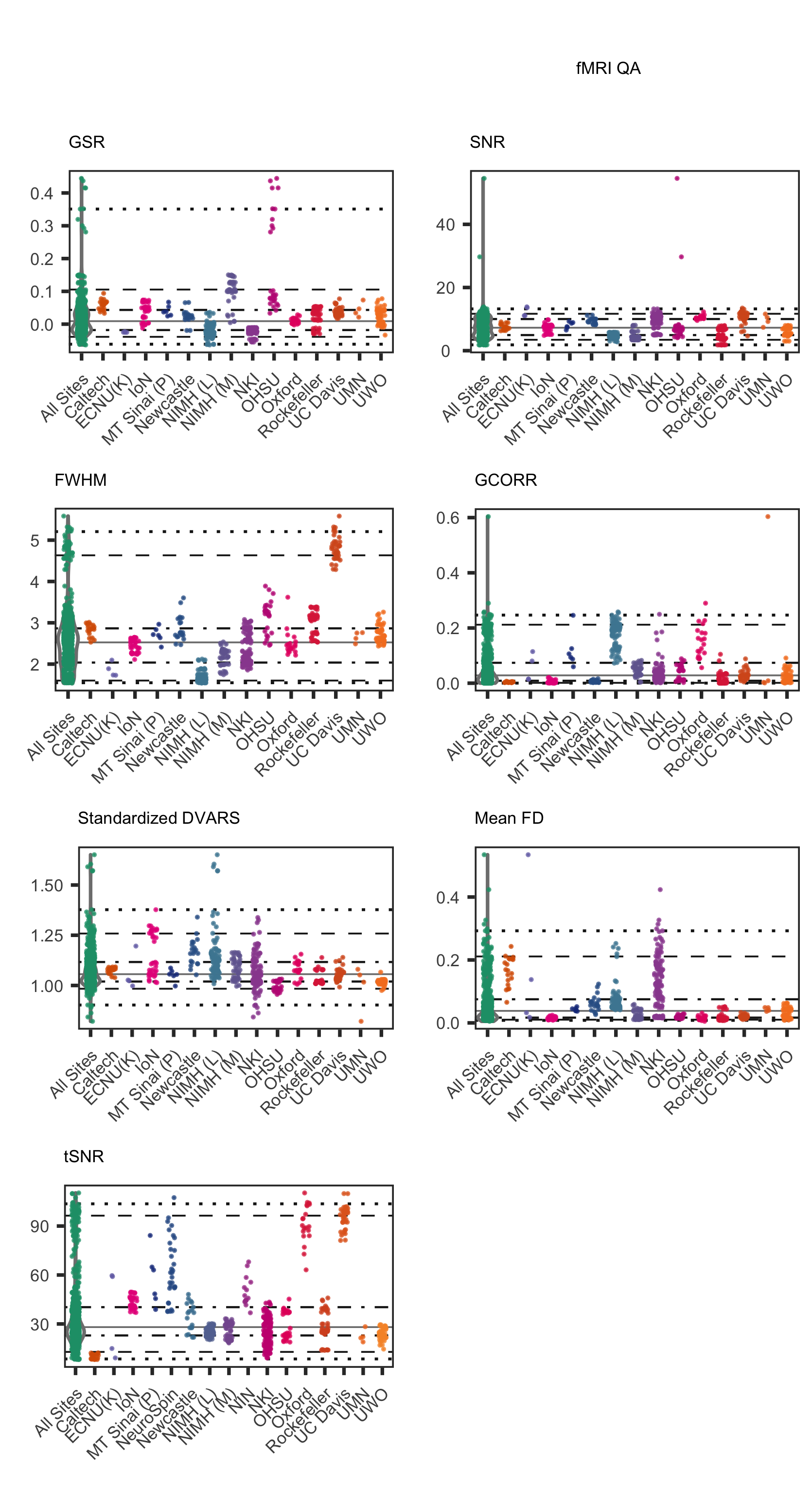
Usage Agreement
Data usage policies differ among the data collections to PRIME depending on the specific institutional and collaborative requirements identified by their contributions. For each collection, the contributor has set the sharing permissions for their data using one or more the following four policies. Usage policies are specified in the Data Collections Summary Table as well as on the individual webpages for the varying collections.
- Creative Commons – Attribution-NonCommercial Share Alike (CC-BY-NC-SA)
Standard INDI data sharing policy. Prohibits use of the data for commercial purposes. - Creative Commons – Attribution (CC-BY)
Least restrictive data sharing policy. - Custom Data Usage Agreement (DUA)
Users must complete a data usage agreement (DUA) prior to gaining access to the data. Contributors can customize the agreement as they see fit, including determining whether or not signatures from authorized institutional official are required prior to executing the DUA.

